Research
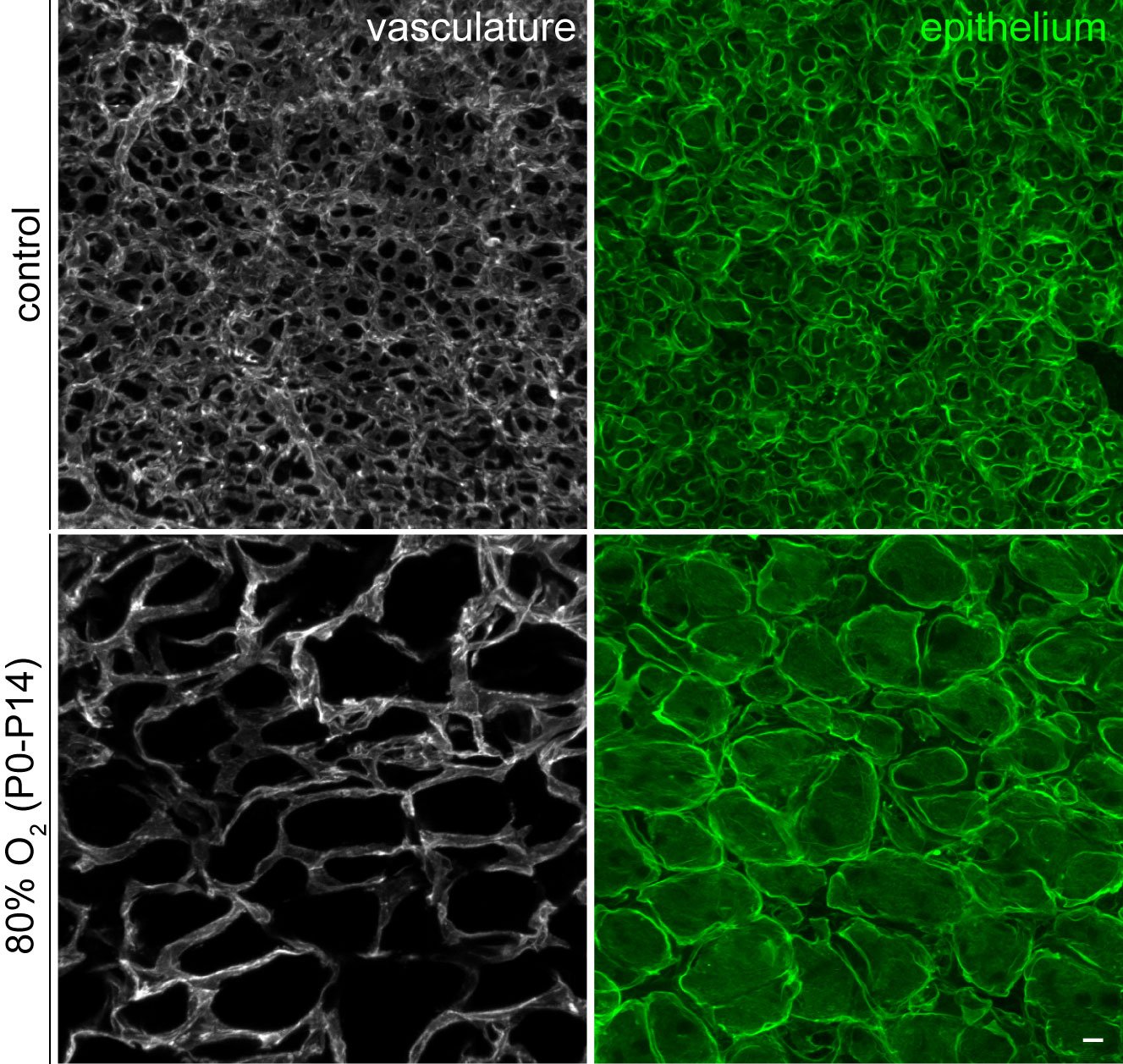
Gas diffusion in the mammalian lung takes place in the alveoli, where a thin epithelium is surrounded by a capillary network. Traditionally, these alveolar capillaries were considered homogeneous until Dr. Vila Ellis' postdoctoral research, which unveiled a novel population of endothelial cells in the mouse lung. Interestingly, the failure to form the alveolar capillary network correctly has been observed in various pulmonary diseases characterized by vascular simplification, such as bronchopulmonary dysplasia (BPD), commonly seen in premature births. Furthermore, recent single-cell RNA sequencing profiling of the human lung by others has also identified a similar endothelial cell population, underscoring the relevance of this research to human diseases.
In the Vila Ellis Lab, our focus lies in investigating this newly discovered heterogeneity during development and disease. Our ultimate objective is to elucidate the transcriptional mechanisms underlying endothelial heterogeneity, endothelial cell fate, and the molecular and cellular components of lung angiogenesis. By doing so, we aim to provide crucial insights into fundamental biology questions.
Mechanism of endothelial cell specification and maturation: The mechanisms underlying lung endothelial fate, maintenance, and recovery, particularly the responsible transcription factors responsible for these processes, remain largely unknown. We used state-of-the-art single-cell ATAC-seq to unbiasedly profile embryonic and postnatal lung endothelial cells, and identified accessible DNA regions specific to individual endothelial populations. We will focus on studying specific candidates derived from our motif enrichment analysis. In order to delve deeper into the role of these transcription factors in endothelial cell fate, we will undertake several approaches. First, we will scrutinize their expression patterns in a cell type-specific manner within the endothelium. Next, we will analyze the effects of their mutation using mouse models, shedding light on their functional significance. Finally, we will conduct ChIP-seq analysis to gain valuable insights into their genomic binding sites and potential downstream targets.
Lung endothelial cell injury-repair in disease models: In the Vila Ellis lab, we investigate how the recently discovered lung endothelial heterogeneity influences pulmonary vascular diseases. Understanding the underlying mechanisms of injury and repair could lead to promising new therapeutic targets, particularly considering its presence in human lungs. Notably, Dr. Vila Ellis’ past research has demonstrated the significance of Car4+ endothelial cells in alveologenesis and their susceptibility to damage in a widely used mouse model of BPD. Additionally, recent publications by others of their involvement in leukocyte trafficking and potential contribution to alveolar regeneration after injury. To gain comprehensive insights, we are employing established mouse lines and state-of-the-art genomics techniques to study cell turnover, fate specification, and regeneration upon viral and hyperoxic injury.
Molecular and cellular mechanisms of intussusceptive angiogenesis: Classical lung morphological studies have associated alveolar angiogenesis with a unique process called intussusceptive angiogenesis (IA), where existing vessels are split by trans-endothelial pillars to form new ones. Despite clear evidence of its existence, IA remains poorly understood. Interestingly, IA features have been recently observed in COVID patients, suggesting a potential pathological or regenerative role after viral infection. Through genetic labeling, we have observed that unlike the filopodia formation seen in sprouting angiogenesis, lung endothelial cells acquire a net-like structure with an attenuated center, indicating the formation of characteristic IA pillars. These observations align with Car4+ endothelial cell morphology, and interestingly, these cells are found in locations where IA is expected to occur. Additionally, both tip cells and Car4+ cells experience Vegfa signaling. Our lab’s goal is to identify the cellular players involved in this process and uncover the molecular components underlying IA, shedding light on this fascinating mechanism.